TARGET PROJECT 1
Gene Editing of the Beta Globin Gene Using TALE Effector Nucleases and ssODNs
Investigator: Dula Man, Ph.D.
Mentors: Eric Kmiec, Ph.D., E. Anders Kolb, M.D.

Dula Man, Ph.D. is an assistant professor at Delaware State University. He obtained his PhD degree in molecular biology from the University of Texas at El Paso, and did x-ray crystallography postdoc at University of California Irvine. Dr. Man has navigate the science fields from molecular biology, biochemistry, structural biology, DNA repair, genome editing to nanomaterial engineering. He published numerous refereed papers in those disciplines. His experience in nanotechnology and biochemistry are indispensable assets to this project.
Project Summary:
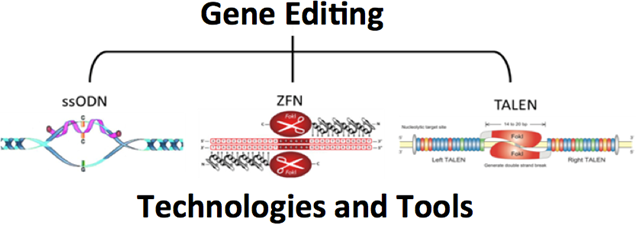
The capacity to reverse an inborn error within the context of the human chromosome has long held a certain fascination for workers in the field of molecular medicine. It is now widely accepted that single-stranded DNA oligonucleotides can direct the genetic exchange of a single base in the DNA and therefore can correct a single base mutation in a disease like Sickle Cell Anemia. This approach is known as gene editing. The mechanism of action and the regulation of gene editing depend on the ability of the ssODN to align in homologous register at the target site in the genome.
While this approach has shown significant scientific merit, the frequency with which gene editing takes place falls short of clinical relevance. Herein we propose to develop a combinatorial approach to gene editing utilizing Transcription Activator-like Effector Nucleases (TALENs) to create a specific double stranded (ds) DNA break site near the mutant base in the chromosome. Ds DNA breaks are known to increase gene editing directed by ssODNs by activating auxiliary DNA repair and cell cycle protein that help catalyze base exchange. We have already demonstrated that the combinatorial activity of ssODNs and TALENs in gene editing enhances the frequency of gene editing by testing in a validated, well established model system with both phenotypic and genotypic readouts.
In this proposal, we intend to apply combinatorial gene editing to the reversal of a βS allele in CD34+ cells. We will characterize and optimize reaction parameters including the length of the ssODN, the cleavage site of TALEN and the position of the mismatch within the paired chromosomal DNA complex. Importantly, we will also evaluate the CD34+ cell for genotoxic and/or metabolic impacts of gene editing process itself. The key is to minimize the downstream effects of that activation by using this new combinatorial approach, while increasing the actual frequency of mutation repair. The aims of this grant are interwoven but also independent and significant information can be gained from the experimental outcomes of each of them. We hope to achieve a working protocol that will advance gene editing toward clinical implementation for Sickle Cell Disease.
Publications:
https://www.ncbi.nlm.nih.gov/myncbi/browse/collection/49592352/?sort=date&direction=ascending
